
Laboratory Investigations in Microbiology

 |
Laboratory Investigations in Microbiology |
 |
As indicated in the last chapter, metabolism encompasses many different chemical reactions of a cell, including those of catabolism (breakdown) and anabolism (build-up). The major difference between bacteria and eukaryotes isn't only in the type of pathways bacteria can use, but in the enormous flexibility and self-sufficiency that many bacteria exhibit. Thus, many bacteria are able to grow with just a single carbon source (e.g. glucose). As long as all the biochemical pathways of a bacterial cell are complete, almost any sugar, fat, or amino acid is sufficient for growth.
Last time we looked at the hydrolysis of macromolecules such as starch and protein. Today we will consider the next steps in metabolism:
Citrate test. Citrate (citric acid) is a
common metabolite of most cells. It is used and produced  during
the Krebs cycle, and most bacteria thus have the enzymes needed to break it
down. However, not every bacterium can use citrate as a source of food. This is
because some bacteria lack the enzyme citrate permease, a transporter
that carries citrate across the plasma membrane into the cytoplasm. We can
determine if a bacterium has this transporter by culturing bacteria on a medium
that contains citrate as the only organic compound. Only bacteria that
can use citrate will grow on this medium. Once inside the cell, citrate is
metabolized as follows: Citrate à
pyruvate + CO2. As citric acid is used up and CO2 diffuses
into the medium (combining with sodium to form sodium carbonate, a base), the pH
of the medium rises, changing the color of the agar from green to blue (AF, KP).
A negative citrate test is inindicated when the medium remains green in color
and bacterial growth fails to take place on the slant (EC).
during
the Krebs cycle, and most bacteria thus have the enzymes needed to break it
down. However, not every bacterium can use citrate as a source of food. This is
because some bacteria lack the enzyme citrate permease, a transporter
that carries citrate across the plasma membrane into the cytoplasm. We can
determine if a bacterium has this transporter by culturing bacteria on a medium
that contains citrate as the only organic compound. Only bacteria that
can use citrate will grow on this medium. Once inside the cell, citrate is
metabolized as follows: Citrate à
pyruvate + CO2. As citric acid is used up and CO2 diffuses
into the medium (combining with sodium to form sodium carbonate, a base), the pH
of the medium rises, changing the color of the agar from green to blue (AF, KP).
A negative citrate test is inindicated when the medium remains green in color
and bacterial growth fails to take place on the slant (EC).
SIM test. Once inside the cell, nutrients are processed to make them "fit" into the cells' metabolic machinery. This machinery usually consists of the biochemical pathways we know as glycolysis, respiration, and fermentation. Not all nutrients are ready to go into these pathways. For example, before amino acids can be used, the cell must remove their nitrogen and sulfur side chains. These reactions are called deamination and desulfurylation, respectively, and can be illustrated by the following examples:
Other side chains found in amino acids are also unusable by the metabolic machinery and must be removed:
These chemical reactions produce measurable quantities of hydrogen sulfid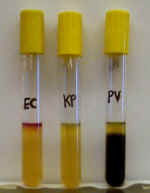 e
gas, indole and ammonium, and the appearance of such waste products indicate
that the bacterium is able to use these amino acids as nutrients. Using a Sulfur-Indole-Motility
(SIM) medium, we can detect the production of two of these compounds.
Hydrogen sulfide gas reacts with iron in the agar to form an insoluble black
precipitate (FeSO4) which is easily visible to the naked eye (PV). In
the absence of H2S, the medium remains yellow (EC, KP). Indole side
chains can be detected by adding a chemical reagent (Kovacs Reagent) to
the top of the agar tube. It reacts with indole to form a pink-red color (EC).
In the absence of indole, Kovacs Reagent remains green (KP, PV).
e
gas, indole and ammonium, and the appearance of such waste products indicate
that the bacterium is able to use these amino acids as nutrients. Using a Sulfur-Indole-Motility
(SIM) medium, we can detect the production of two of these compounds.
Hydrogen sulfide gas reacts with iron in the agar to form an insoluble black
precipitate (FeSO4) which is easily visible to the naked eye (PV). In
the absence of H2S, the medium remains yellow (EC, KP). Indole side
chains can be detected by adding a chemical reagent (Kovacs Reagent) to
the top of the agar tube. It reacts with indole to form a pink-red color (EC).
In the absence of indole, Kovacs Reagent remains green (KP, PV).
Oxidase test and Nitrate reduction test. When bacteria carry out respiration, nutrients such as glucose or amino acids are completely "burned" to carbon dioxide. The electrons and hydrogen gained from these metabolites are carried to the electron transport chain, which in turn converts their energy into ATP usable by the cell. At the end of the ETC, a final electron acceptor combines with the electrons (and often, H) to form a waste product which is then lost from the cell. The waste product generated depends on the type of respiration carried out:
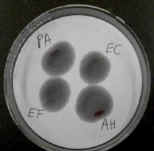
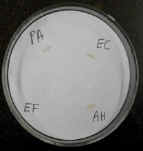 The oxidase test uses a reagent that is reduced by cytochrome oxidase.
Bacterial colonies that have this enzyme turn purple or black.
The oxidase test uses a reagent that is reduced by cytochrome oxidase.
Bacterial colonies that have this enzyme turn purple or black.
The nitrate reduction test detects the presence of nitrite in the medium. Nitrite is only produced by bacteria that carry out anaerobic respiration using nitrate. Nitrite can be detected by adding two test reagents (A and B) to the medium. If nitrite was produced, the medium turns pink/red within 5 minutes (often faster). This indicates a positive test for nitrate reduction (EC). If nitrate was not reduced the color of the medium will not change color, indicating a possible negative test result.
 However,
some bacteria, notably strains of Pseudomonas, carry out a process called
denitrification. Instead of reducing nitrate to nitrite, these bacteria
continue to add electrons to nitrite, eventually forming N2 gas.
Since nitrite is therefore not detected in the medium, the test appears
to be negative. We can check whether denitrification has taken place by
performing a zinc test. Adding zinc to a medium containing nitrate will
change it to nitrite. A red color appears since the nitrite now reacts with the
reagents (A + B). This confirms a negative test for nitrate reduction
(AF). However, if a bacterium has carried out denitrification, no nitrate is
available to be changed by the zinc, and adding zinc will not produce a
red color. This indicates a positive test for denitrification (SOIL).
However,
some bacteria, notably strains of Pseudomonas, carry out a process called
denitrification. Instead of reducing nitrate to nitrite, these bacteria
continue to add electrons to nitrite, eventually forming N2 gas.
Since nitrite is therefore not detected in the medium, the test appears
to be negative. We can check whether denitrification has taken place by
performing a zinc test. Adding zinc to a medium containing nitrate will
change it to nitrite. A red color appears since the nitrite now reacts with the
reagents (A + B). This confirms a negative test for nitrate reduction
(AF). However, if a bacterium has carried out denitrification, no nitrate is
available to be changed by the zinc, and adding zinc will not produce a
red color. This indicates a positive test for denitrification (SOIL).
Next class:
Next class:
© 2003 - 2019 José de Ondarza, Ph.D.